Frequently Asked Questions
Cropped field-of-view
Some issues on this: #273, #207, #125.
Why does this happen? suite2p crops the field-of-view so that areas that move out of view on the edges are not used for ROI detection and signal extraction. These areas are excluded because they are not always in the FOV - they move in and out and therefore activity in these regions is unreliable to estimate.
suite2p determines the region to crop based on the maximum rigid shifts in XY. You can view these shifts with the movie in the “View registered binary” window. If these shifts are too large and don’t seem to be accurate (low SNR regime), you can decrease the maximum shift that suite2p can estimate by setting ops[‘maxregshift’] lower than its default (which is 0.1 = 10% of the size of the FOV). suite2p does exclude some of the large outlier shifts when computing the crop, and determines the threshold of what is an “outlier” using the parameter ops[‘th_badframes’]. Set this lower to increase the number of “outliers”. These “outliers” are labelled as ops[‘badframes’] and these frames are excluded also from ROI detection.
You can add frames to this list of ops[‘badframes’] by creating a numpy array (0-based, the first frame is zero) and save it as bad_frames.npy in the folder with your tiffs (if you have multiple folders, save it in the FIRST folder with tiffs, or if you have subfolders with ‘look_one_level_down’ it should be in the parent folder). See this page inputs for more info.
Deconvolution means what?
There is a lot of misinformation about what deconvolution is and what it isn’t. Some issues on this: #267, #202, #169
TLDR: Deconvolution will NOT tell you how many spikes happened in a neuron - there is too much variability in the calcium signal to know that. Our deconvolution has NO sparsity constraints and we recommend against thresholding the output values because they contain information about approximately how many spikes occurred. We found that using the raw deconvolved values gave us the most reliable responses to stimuli (as measured by signal variance).
See this figure from our review paper for reference:
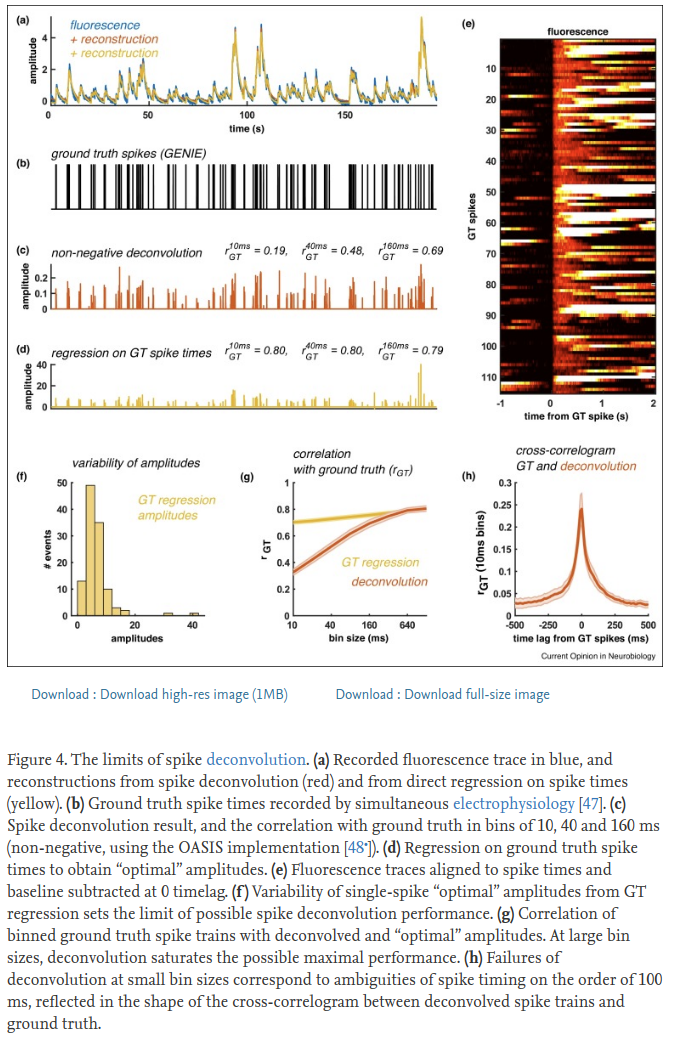
Long answer (mostly from #267):
There is an unknown scaling factor between fluorescence and # spikes, which is very hard to estimate. This is true both for the raw dF, or dF/F, and for the deconvolved amplitudes, which we usually treat as arbitrary units. The same calcium amplitude transient may have been generated by a single spike, or by a burst of many spikes, and for many neurons it is very hard to disentangle these, so we don’t try. Few spike deconvolution algorithms try to estimate single spike amplitude (look up “MLspike”), but we are in general suspicious of the results, and usually have no need for absolute numbers of spikes.
As for the question of thresholding, we always recommend not to, because you will lose information. More importantly, you will treat 1-spike events the same as 10-spike events, which isn’t right. There are several L0-based methods that return discrete spike times, including one we’ve developed in the past, which we’ve since shown to be worse than the vanilla OASIS method (see our Jneurosci paper). We do not use L1 penalties either, departing from the original OASIS paper, because we found that hurts in all cases (see Jneurosci).
How do you compare across cells then if these values are arbitrary to some extent?
If you need to compare between cells, you would usually be comparing effect sizes, such as tuning width, SNR, choice index etc. which are relative quantities, i.e. firing rate 1 / firing rate 2. If you really need to compare absolute firing rates, then you need to normalize the deconvolved events by the F0 of the fluorescence trace, because the dF/F should be more closely related to absolute firing rate. Computing the F0 has problems in itself, as it may sometimes be estimated to be negative or near-zero for high SNR sensors like gcamp6 and 7. You could take the mean F0 before subtracting the neuropil and normalize by that, and then decide on a threshold to use across all cells, but at that point you need to realize these choices will affect your result and interpretation, so you cannot really put much weight on them. For these reasons, I would avoid making statements about absolute firing rates from calcium imaging data, and I don’t know of many papers that make such statements.
Multiple functional channels
If you have two channels and they both have functional activity, then to process both you need to run suite2p in a jupyter notebook. Here is an example notebook for that purpose: multiple_functional_channels.ipynb
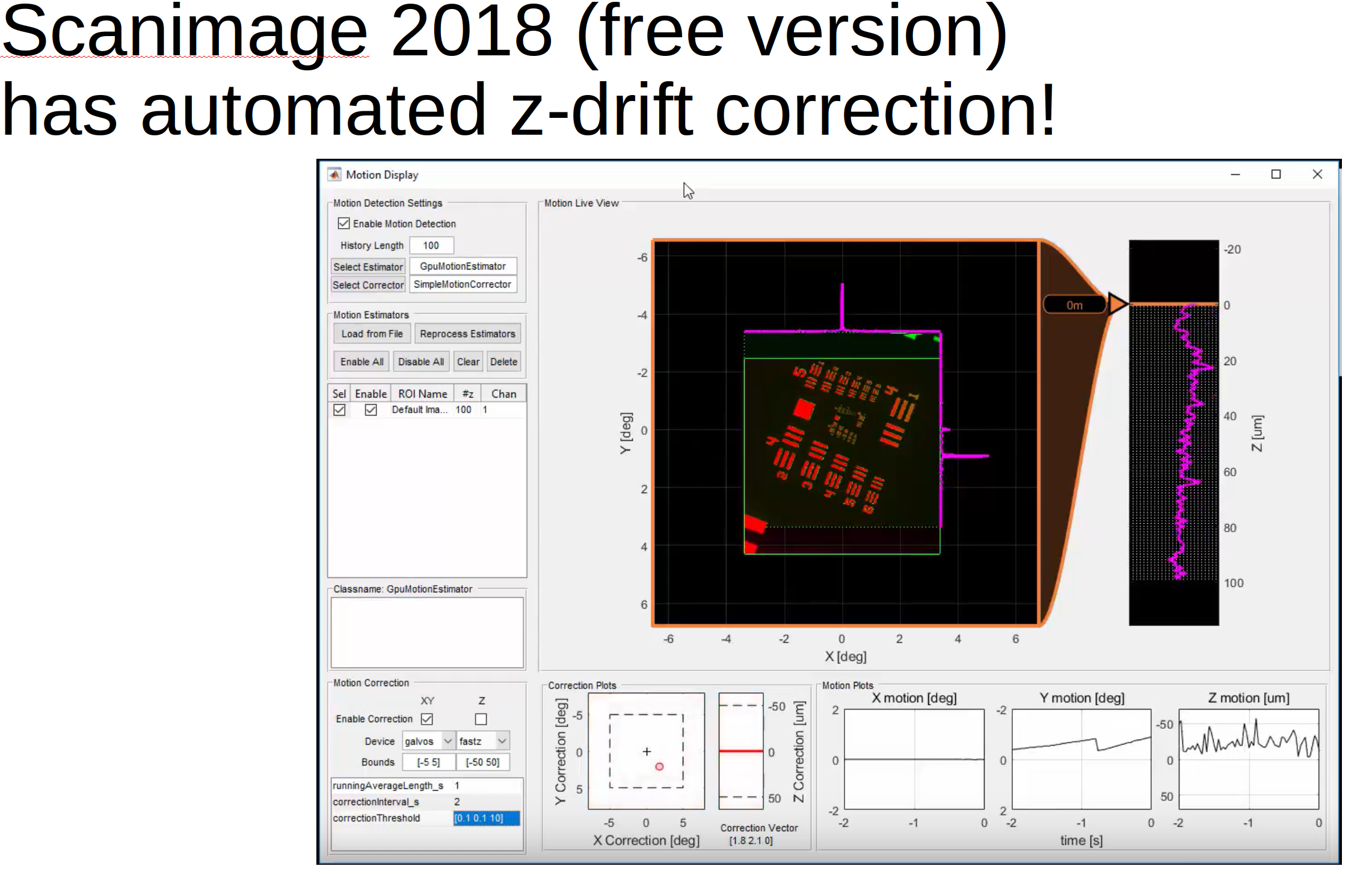
Z-drift
It’s not frequently asked about but it should be :)
In the GUI in the “View registered binary” window you can now load in a z-stack and compute the z-position of the recording across time.
Scanimage now can do z-correction ONLINE for you!
No signals in manually selected ROIs
If you circle an ROI in the manual selection GUI on top of another ROI and ops[‘allow_overlap’] is 0 or False, then that ROI will have no activity because it has no non-overlapping pixels. You can change this after processing with
import numpy as np
np.load('ops.npy', allow_pickle=True).item()
np.save('ops_original.npy', ops)
ops['allow_overlap'] = True
np.save('ops.npy', ops)
Thanks @marysethomas, see full issue here: #651,